Example Pipeline¶
Let us consider a bioinformatics pipeline with four stages as shown in the following figure:
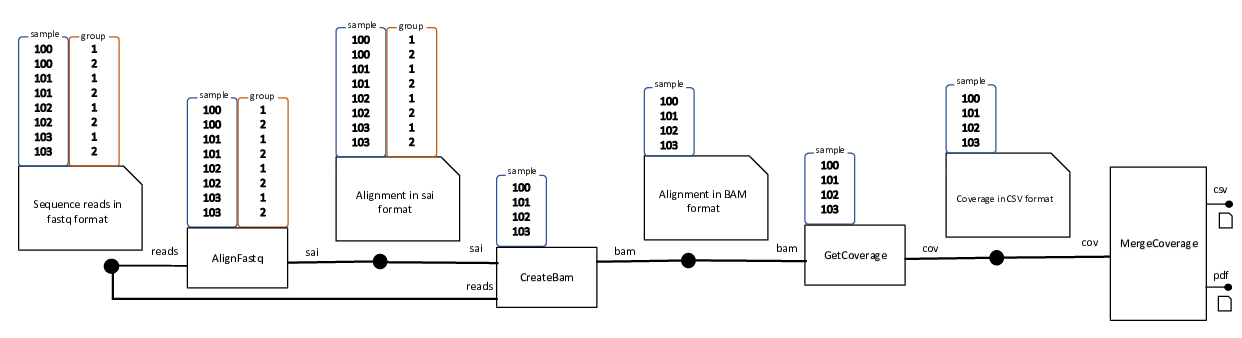
In the first stage each of the 8 input FASTQ file of the form {sample}_{group}.fq, where
sample can be any of 100,101,102,103 and group can be 1 or 2, is aligned to a reference genome
and creates a temporary alignment file.
In the second stage, for each sample the two temporary alignment files for the two groups are converted to a BAM alignment file.
In the third stage, a genome coverage table is generated for each sample.
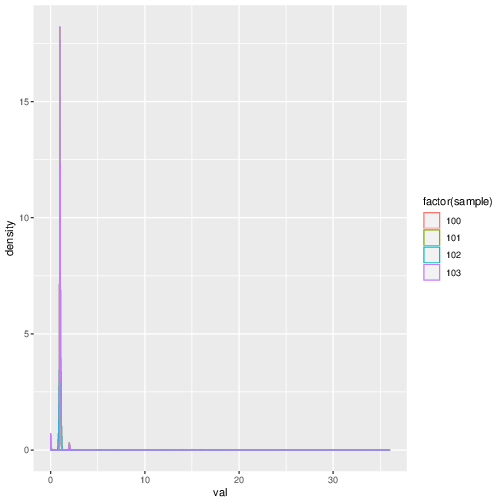
In the final stage, the coverage tables for the four samples are consolidated into a single coverage table. A a plot containing the coverage densities of the four samples as shown below is also created.